Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Matsumoto, Atsushi
no journal, ,
Kanaeda, Naoko; Ishida, Hisashi; Kono, Hidetoshi
no journal, ,
The nucleosome, consisting of a histone octomer with DNA wrapped around it, is the basic repeating unit of the eukaryotic chromosome structure. Because the DNA is wrapped around the histone core, it is difficult for proteins that regulate transcription, DNA replication, repair and recombination to access the nucleosomal DNA. Nucleosomes have been known to dynamically change their positions in accordance with cell states. ATP-hydrolyzing chromatin remodelers are considered to loosen the form of the DNA and relocate the DNA inside the nucleosome in order to expose cis-elements to which regulatory proteins bind. We believe that the free energy profile of nucleosomal DNA sliding can provide some important clues to help understand the mechanism of changes in nucleosome positioning.
 sp.593 (HaNDK)
sp.593 (HaNDK)Arai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
The nucleoside diphosphate kinases (NDKs) are known to have a tetrameric or hexameric oligomer structure formed by association of common dimeric components. In order to understand the oligomeric structure of NDK, the wild-type NDK from Halomonas sp. 593 (HaNDK) was determined by X-ray crystallography. The wild-type HaNDK only exhibited a dimeric form that is the same as a common dimer unit seen in other NDKs. This is the first evidence of a dimeric structure for HaNDK. To explore the effect of mutation on the assembly form of HaNDK, E134, which is a key interface residue for other tetrameric NDKs, was replaced with Ala, and the structure was determined by X-ray crystallography. Two kinds of tetrameric assemblies, Type I and Type II, appeared in the asymmetric unit of the E134A mutant HaNDK. The change from dimeric to tetrameric assembly is attributed to the removal of negative charge repulsion caused by the E134 in the wild-type HaNDK.
Yonetani, Yoshiteru; Kono, Hidetoshi
no journal, ,
Yonetani, Yoshiteru; Kono, Hidetoshi
no journal, ,
Higuchi, Mariko; Pinak, M.
no journal, ,
Ionizing radiation leads to clustered DNA damage sites, which are defined as two or more single lesions induced within 10-20 base pairs (bp). The detail mechanism of repair of multiple lesions is not known. The experimental results showed that the excision of the 8-oxo-7,8-dihydroguanine (8oxoG) lesion by repair enzyme hOGG1 is strongly inhibited by the presence of a single strand break (SSB) on the opposite strand with a 1bp separation from the 8oxoG lesion and probability of repair increases with increasing distance between the SSB and the 8oxoG. In this study, we present results of molecular dynamics simulation of the models of complex of hOGG1 and cluster damaged DNA. Each model DNA contains two damages, SSB and 8oxoG. According to the observed results, the structural fluctuation of cluster damaged DNA decreased largely when hOGG1 bound the cluster DNA. The effect of the SSB located at -3bp was small because the residue located at -3bp did not contact directly with hOGG1.
Murakami, Hiroshi; Ono, Masato
no journal, ,
no abstracts in English
Nakagawa, Hiroshi
no journal, ,
Protein is a functional element in all living organisms. Almost all vital phenomena are mediated by specific proteins. Proteins work in an aqueous environment at ambient temperature, indicating that proteins cannot escape from thermal fluctuations. The magnitude of physiologically relevant input is as the same level as thermal fluctuations. It is widely accepted that the proteins are flexible and mobile. The flexibility and mobility, that is, protein dynamics are essential for protein functions. Understanding of protein dynamics is also important to clarify how the proteins can discriminate physiologically relevant motions from random fluctuations. Neutron incoherent scattering is one of the most powerful techniques to observe protein dynamics quantitatively, for example, vibration and diffusion dynamics. The next-generation neutron source, J-PARC, in Ibaraki was constructed and a several neutron instruments are already working. We expect that the neutron source will open new era for biophysics. Here I will talk about dynamics of protein and its hydration water, and discuss the prospect of the dynamics study of biomolecules at the J-PARC.
Akamatsu, Ken; Shikazono, Naoya
no journal, ,
Clustered damage site is believed to hardly be repaired. This type of damage is considered to be induced around high-LET ionizing radiation tracks and at track-end of secondary electron. However, details of the clustered damage sites are not known. In order to get experimental information about the hypothetical damage, we have developed an analytical method for measuring the degree of dispersion of distances between DNA lesions using fluorescence resonance energy transfer (FRET). Complementary two single-strand DNA with an AP-site at each center nucleotide were synthesized to obtain relationship between FRET efficiency and AP-site- AP-site distance. As a result of fluorescence spectral analyses with annealing these strands, fluorescence intensity from acceptor increased with proceeding the annealing. D-A distance calculated from the FRET efficiency was 4 nm, which was reasonable with the estimated one taking the diameter of B-form DNA duplex and length of each probe into account.
 2 chain
2 chainMatsumoto, Fumiko; Hatanaka, Takaaki*; Tamada, Taro; Honjo, Eijiro*; Ota, Shoichiro*; Ito, Yuji*; Izuhara, Kenji*; Kuroki, Ryota
no journal, ,
Interleukin-13 (IL-13) is a key cytokine critical to the development of T-cell-mediated humoral immune responses which are associated with allergy and asthma. IL-13 possesses two types of receptor: the heterodimer, composed of IL-13R 1 and IL-4R
1 and IL-4R , transducing the IL-13 signals; and the IL-13R
, transducing the IL-13 signals; and the IL-13R 2, it has high affinity rather than IL-13R
2, it has high affinity rather than IL-13R 1 against IL-13, acting as a nonsignaling "decoy" receptor. In order to investigate the inhibition mechanism of IL-13 signal by IL-13R
1 against IL-13, acting as a nonsignaling "decoy" receptor. In order to investigate the inhibition mechanism of IL-13 signal by IL-13R 2, extra cellular region of IL-13R
2, extra cellular region of IL-13R 2 was expressed by silkworm-baculovirus expression system after fusing the DNA cording the extracellular region of human IL-13R
2 was expressed by silkworm-baculovirus expression system after fusing the DNA cording the extracellular region of human IL-13R 2 with the Fc derived from mouse IgG2a. The expressed fusion protein (IL-13R
2 with the Fc derived from mouse IgG2a. The expressed fusion protein (IL-13R 2)2-Fc was purified by a protein A column followed by an ion exchange column. The affinities of the ligand complexes, IL-13/IL-13R
2)2-Fc was purified by a protein A column followed by an ion exchange column. The affinities of the ligand complexes, IL-13/IL-13R 1 and IL-13/IL-13R
1 and IL-13/IL-13R 2 to (IL-4R
2 to (IL-4R )2-Fc were investigated to explore the inactivation mechanism of the IL-13R
)2-Fc were investigated to explore the inactivation mechanism of the IL-13R 1 signal with IL-13R
1 signal with IL-13R 2 by surface plasmon resonance analysis. IL-13/IL-13R
2 by surface plasmon resonance analysis. IL-13/IL-13R 1 complex has an affinity (KD=1.20
1 complex has an affinity (KD=1.20 10
10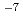 M) with (IL-4Ra)2-Fc, whereas the IL-13/IL-13R
M) with (IL-4Ra)2-Fc, whereas the IL-13/IL-13R 2 complex had no affinity with (IL-4R
2 complex had no affinity with (IL-4R )2-Fc. This observation suggests that IL-13R
)2-Fc. This observation suggests that IL-13R 2 does not inactivate the IL-13R
2 does not inactivate the IL-13R 1 through formation of a ternary complex with IL-13 and IL-4R
1 through formation of a ternary complex with IL-13 and IL-4R , but removes the IL-13 with its higher affinity to IL-13 without forming a ternary complex.
, but removes the IL-13 with its higher affinity to IL-13 without forming a ternary complex.
Masui, Tomomi; Fujiwara, Satoru; Nakagawa, Hiroshi; Kataoka, Mikio
no journal, ,
It is known that trehalose is a protective reagent and plays important roles in desiccation tolerance. One of the important properties of trehalose for the desiccation tolerance is stabilization of the biomembranes in the fluid phase. However, due to lack of information of how the trehalose molecules distribute on the biomembrane and interact with lipid molecules, the preservation mechanism of the biomembrane by trehalose has not been elucidated. Here we aimed at determining the distribution of trehalose on the simple model biomembrane and characterize dynamic properties of the membrane and the lipid molecules. For this purpose we employed small- and wide-angle X-ray scattering (SAXS and WAXS) and neutron spin echo (NSE) techniques. The NSE measurement revealed that the addition of trehalose to lipid membrane decreases the bending rigidity. This effect of trehalose is same tendency of temperature.
Fujiwara, Satoru; Yonezawa, Yasushige*; Matsumoto, Fumiko; Oda, Toshiro*; Takeda, Soichi*
no journal, ,
no abstracts in English
Kanaeda, Naoko; Ishida, Hisashi; Kono, Hidetoshi
no journal, ,
no abstracts in English
Tokuhisa, Atsushi*; Kai, Takeshi; Taka, Junichiro*; Kono, Hidetoshi; Go, Nobuhiro*
no journal, ,